Experimental design
Coverage of the surveyed retail outlets represented all major geographical regions and price tiers in Hong Kong. Sample collection was performed between November 2017 and February 2018. At every retail outlet surveyed, one of each eel product available was purchased and processed for identification, even when the same products were stocked across different stores. In general, Hong Kong supermarkets display considerable homogeneity in stocked products, particularly in convenience stores (vendors A6 and A7) for which the eel products purchased were all of the in-house private label brand.
Samples were composed of raw, frozen, previously cooked, and ready-to-eat (sushi) eel meat. DNA was extracted from 2 to 3 mm3 pieces of tissue based on the QIAGEN DNeasy kit (QIAGEN Inc.) manufacturer’s protocol, along with a negative extraction control. All extractions were run on a 1.5% agarose gel and met visual quality standards, i.e., a bright, high–molecular weight band with little to no smearing. We then targeted a 655–base pair fragment of the mitochondrial cytochrome oxidase subunit I (COI) for amplification using polymerase chain reaction (PCR) using primers proven to be effective for fish species amplification and identification (38).
FishF1-5′ TCA ACC AAC CAC AAA GAC ATT GGC AC 3′
FishR1-5′ TAG ACT TCT GGG TGG CCA AAG AAT CA 3′
Each 20-μl reaction contained 10 μl of Biotech Rabbit 2× Hotstart Lyophilized PCR Mastermix (Biotech Rabbit, Germany), 1 μM of each primer, and 2 μl of template DNA. PCR was carried out on a Veriti 96-Well Thermal Cycler (Applied Biosystems Inc.) with the following program: initial activation at 95°C for 2 min, followed by 35 cycles of denaturing at 95°C for 30 s, annealing at 48° for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 5 min. A no template control and a positive control containing DNA extracted from another fish species that had previously been amplified and sequenced successfully were also used.
PCR products were cycle sequenced using the BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems Inc.). Each cycle sequencing reaction consisted of 4.0 μl of BigDye, 3 μl of PCR product, 3 μl of ultrapure ddH2O, and 1 μl of primer (3.2 pmol; COI FishF1 or COI FishR1). Sequencing was performed on an Applied Biosystems Genetic Analyzer 3130xl. Sequence chromatograms were assessed visually, and consensus sequences were generated in Geneious v10.0.7. Considering the closely related nature of the species in question (A. anguilla, A. japonica, and A. rostrata), the species identity of each sample was tested in three ways further detailed below: (i) comparison to entries within GenBank database using the BLAST algorithm and (ii) BOLD database using the BOLD ID engine, and (iii) phylogenetic tree building and species cluster delineations (see table S1) (39, 40).
First, consensus sequences for each COI barcoding region were compared to the GenBank sequence database for preliminary identification. To prevent potential misidentifications inherent in open source databases, the best matched sequence from GenBank was checked within BOLD, in which entries are more stringently validated. Several GenBank accessions did not match their BOLD IDs and so were not considered. We also compared sequences generated in this study directly within the BOLD database.
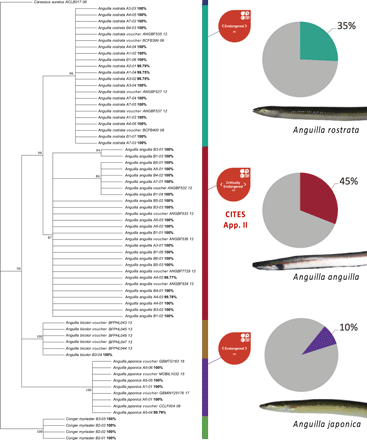
Last, a phylogenetic tree (Fig. 1) was constructed using reference sequences validated as mentioned above (BOLD and GenBank in agreement), and sequences from this study were mapped against it. The tree includes sequences generated in this study using COI primers and a subset validated and publicly available on the BOLD (www.barcodinglife.org) with accession numbers indicated. Sequences were aligned in MEGA7 (39) using the ClustalW algorithm, manually checked, and adjusted for a frameshift error. Model selection was also performed in MEGA7 with Tamura 3-parameter+I model yielding the lowest Bayesian Information Criterion score. The maximum-likelihood tree was constructed in MEGA7 using 1000 bootstrap replicates, a cutoff value of 85%, and support values indicated at each node. Carassius auratus was used as the outgroup.
Photos provided by Ryoshiro Wakiya, Florian M. Stein, and Erling Svensen (WWF).